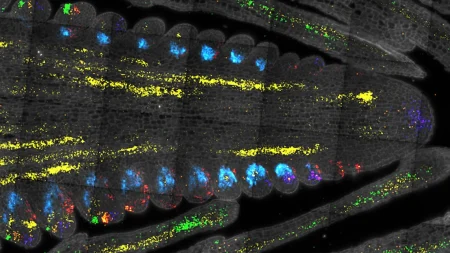
In a recent study by researchers from Carnegie Mellon University’s School of Computer Science, a new machine learning method called scGHOST has been developed to detect subcompartments in the 3D genome of human cells. Jian Ma, along with former Ph.D. students Kyle Xiong and Ruochi Zhang, introduced this method in a paper published by the journal Nature Methods. These subcompartments provide insights into the spatial organization of chromosomes within the nucleus and how they influence gene expression patterns.
The organization of the human genome in 3D structures is crucial for understanding how DNA structure impacts gene expression and disease processes. By exploring the connections between cellular structure and function, researchers aim to uncover the relationships between chromosome organization and gene expression. The development of scGHOST addresses the challenge of poor data quality when studying these structures at the single-cell level. Using graph-based machine learning, scGHOST enhances data quality, making it easier to identify and locate how chromosomes are spatially arranged.
By accurately identifying 3D genome subcompartments, scGHOST contributes to the growing collection of single-cell analysis tools used by scientists to study the complex molecular landscape of tissues, particularly in the brain. This advancement in understanding the cellular structure and gene regulation at a single-cell level holds promise for uncovering new insights into health and disease processes. With the ability to connect chromosome organization within the nucleus to gene expression patterns, researchers can gain a deeper understanding of how DNA structure influences biological functions.
The ultimate goal of single-cell biology is to elucidate the relationships between cellular structure and function in various biological contexts. By studying the 3D genome structures within the cell nucleus, researchers can uncover valuable insights into how genes are regulated and how this regulation affects different biological processes. The scGHOST method offers a novel approach to studying the spatial organization of chromosomes within the nucleus, providing researchers with a powerful tool to analyze gene expression patterns and their relationship to chromosome organization.
The advancements in technology that allow for the study of 3D genome structures at the single-cell level have revolutionized the field of genetics and molecular biology. By analyzing the organization of chromosomes within the nucleus, researchers can better understand how gene expression is controlled and how this process is influenced by the spatial arrangement of DNA. The development of scGHOST represents a significant leap forward in our ability to study the intricate molecular landscape of complex tissues, offering new opportunities to explore gene regulation in health and disease contexts.
As researchers continue to explore the connections between cellular structure, chromosome organization, and gene expression patterns, new discoveries are being made that have the potential to transform our understanding of human genetics and disease processes. The development of scGHOST represents a major advancement in the field of single-cell biology, providing scientists with a powerful tool to study the spatial organization of chromosomes within the nucleus and its impact on gene expression. With the ability to accurately detect and analyze 3D genome subcompartments, scGHOST opens up new avenues for research into gene regulation and disease mechanisms.