Summarize this content to 2000 words in 6 paragraphs Researchers from Children’s Hospital of Philadelphia (CHOP) and Stanford University have revealed the molecular structure of TRACeR-I, a protein platform for reprogramming immune responses. A better understanding of its structure may help optimize designs for the platform, which can be used to develop cancer treatments by either directly modifying immune cells or by creating proteins that help immune cells locate cancer cells. The findings were published today by the journal Nature Biotechnology.
Immunotherapy presents a promising strategy for treating cancer, autoimmune diseases and viral infections, but its effectiveness depends on its ability to specifically target diseases cells. Monoclonal antibodies are widely used because they can target antigens — proteins generated by cancer cells that trigger an immune response — on the surface of diseased cells, but uniquely expressed antigens found on the surface are sparse.
Another potentially powerful target involves fragments of these proteins may be presented on the tumor cell surface through the presentation of peptides on the major histocompatibility complex (MHC), which displays pieces of suspicious material like parts of a virus or cancer cells on the surface of our cells. There are more than 30,000 different versions of MHC-I proteins in humans, which makes it incredibly challenging to develop treatments that can recognize these peptides across large groups of patients and treat a variety of diseases.
Researchers at Stanford make a breakthrough with the development of TRACeRs, platforms that recognizes many different versions of these MHC proteins. TRACeRs act as “master keys” that can open a variety of “locks” posed by these MHC proteins and then treat the appropriate diseased cells while sparing healthy cells.
“Our TRACeR-I and TRACeR-II platforms unlock the potential for targeting disease-associated class I and class II MHC antigens through novel binding mechanisms that overcomes many of the hurdles that have historically limited the broader development of MHC-targeting molecules,” said senior author Possu Huang, PhD, an assistant professor in the Department of Bioengineering at Stanford University. “Our platforms have high peptide-focused specificity, broad compatibility with a variety of antigens and simpler development that significantly expand the accessibility of targetable MHC biomarkers.”
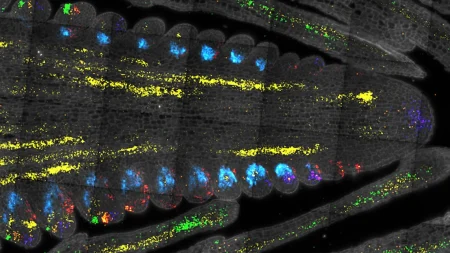
To better understand the potential of the TRACeR-I platform, researchers from CHOP used x-ray crystallography to show exactly how the platform attaches to parts of the MHC-I complex that stay the same across different versions while continuing to recognize the peptides that indicate cancer cells or other dangerous material being displayed on the surface.
“We revealed TRACeR-I’s novel binding mechanism and how the structure of this platform is able to help it recognize surface proteins that indicate cancer cells,” said Nikolaos Sgourakis, PhD, Associate Professor in the Center for Computational and Genomic Medicine at CHOP. “With this collaborative work, we were able to take the Huang lab’s designs and help realizing their exciting therapeutic potential.”
This study was supported by a Stanford Bio-X Graduate Fellowship, a Stanford Graduate Fellowship Award, the NIH Biotechnology Training Program, the NIH Biophysics Training Program, The Mark Foundation for Cancer Research, the Postdoc Mobility fellowship from the Swiss National Science Foundation, the American Cancer Society grant ACS134055- 441 IRG-218, Stanford School of Medicine, Discovery Innovation Fund, NIH grants 442 R01 AI143997, R35 GM125034, U01 DK112217, the Children’s Hospital of Philadelphia Cell and Gene Therapy Collaborative, and the Asplundh foundation.
Keep Reading
Subscribe to Updates
Get the latest creative news from FooBar about art, design and business.
© 2025 Globe Timeline. All Rights Reserved.